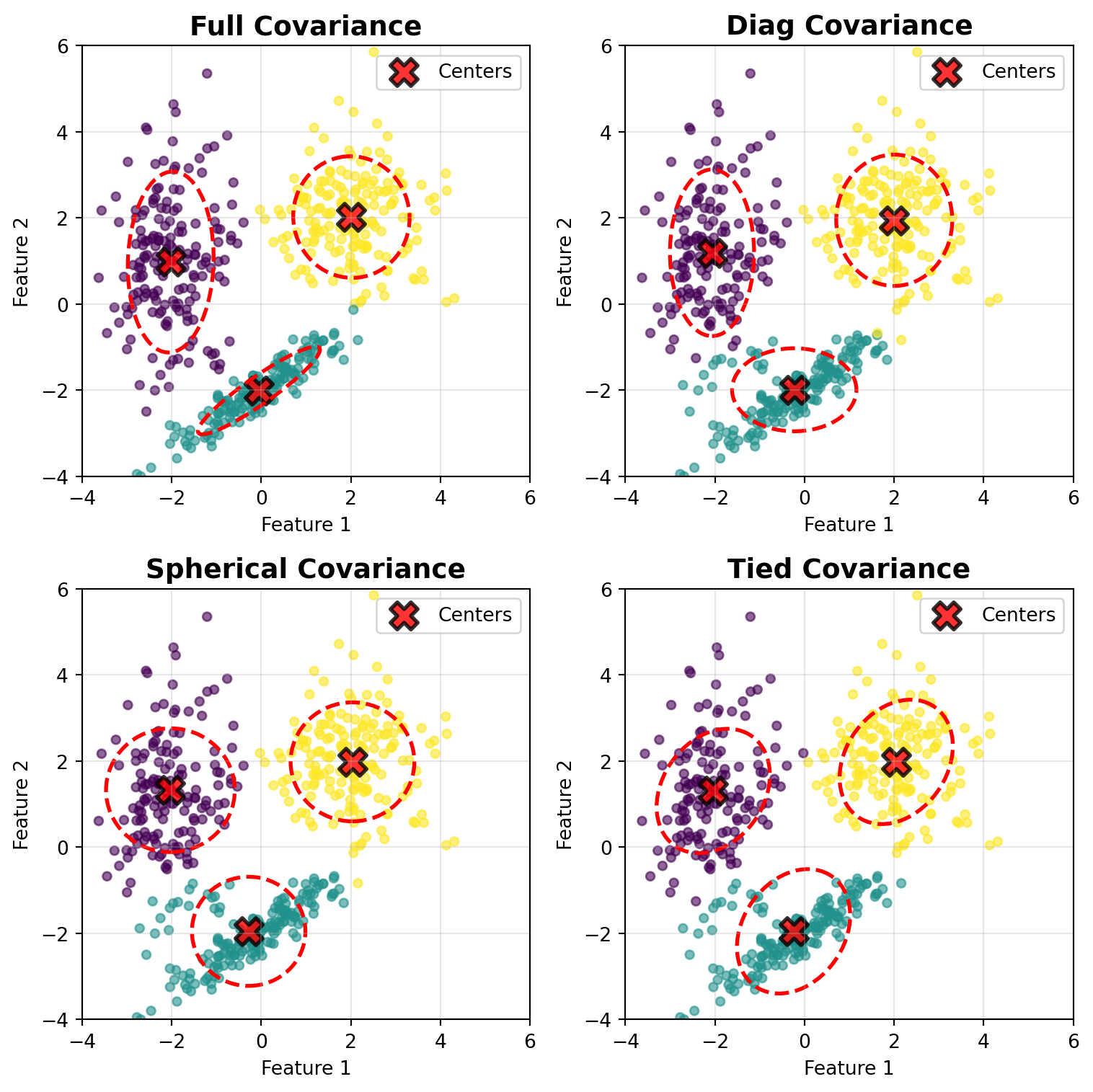
from sklearn.mixture import GaussianMixture
from sklearn.datasets import make_blobs
# Generate synthetic data with 3 clusters using different covariance matrices
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
# Create data with different covariance structures
np.random.seed(42)
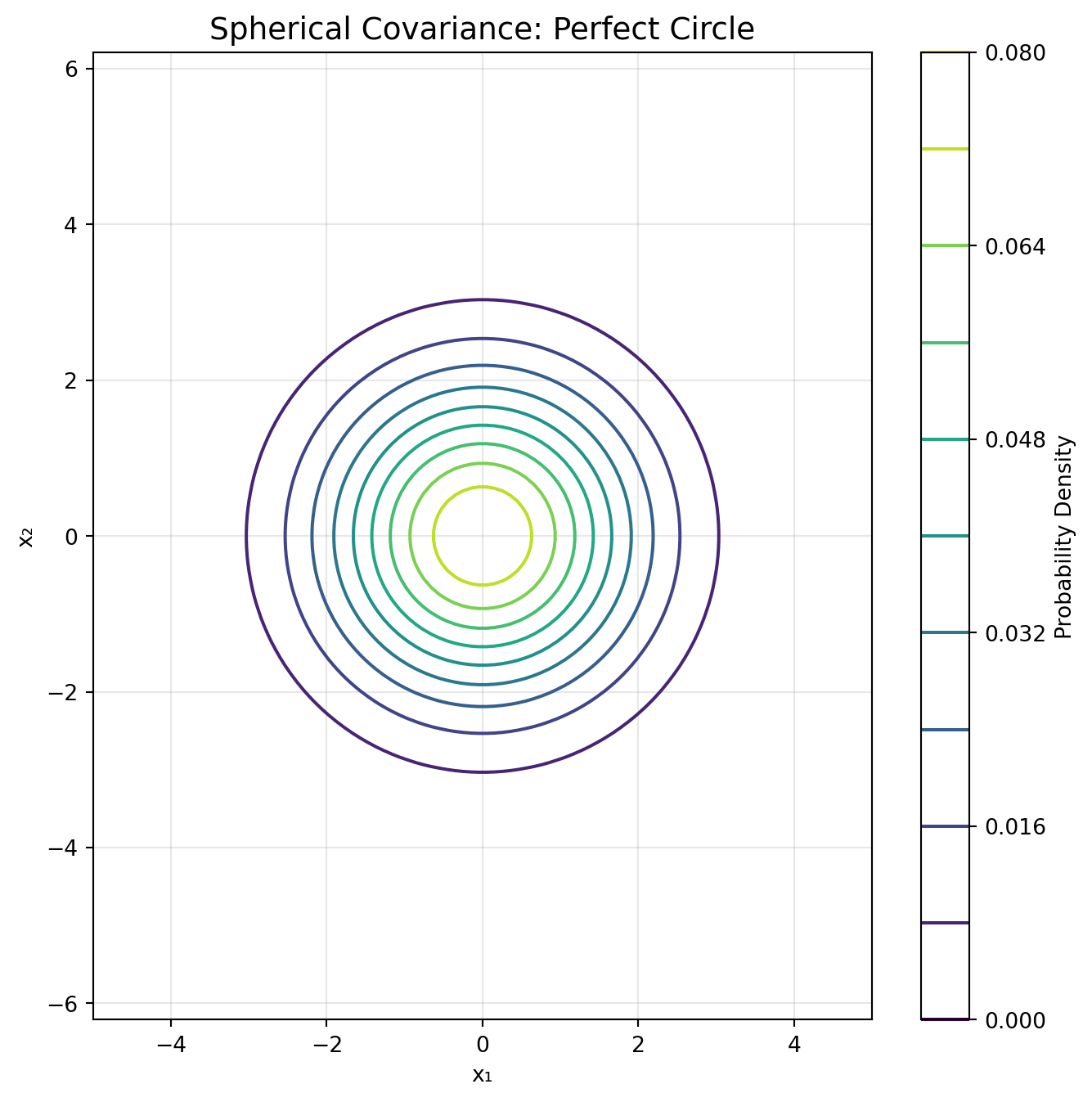
# Cluster 1: Spherical (circular)
n1 = 167
X1 = np.random.multivariate_normal([2, 2], [[1.0, 0], [0, 1.0]], n1)
y1 = np.zeros(n1)
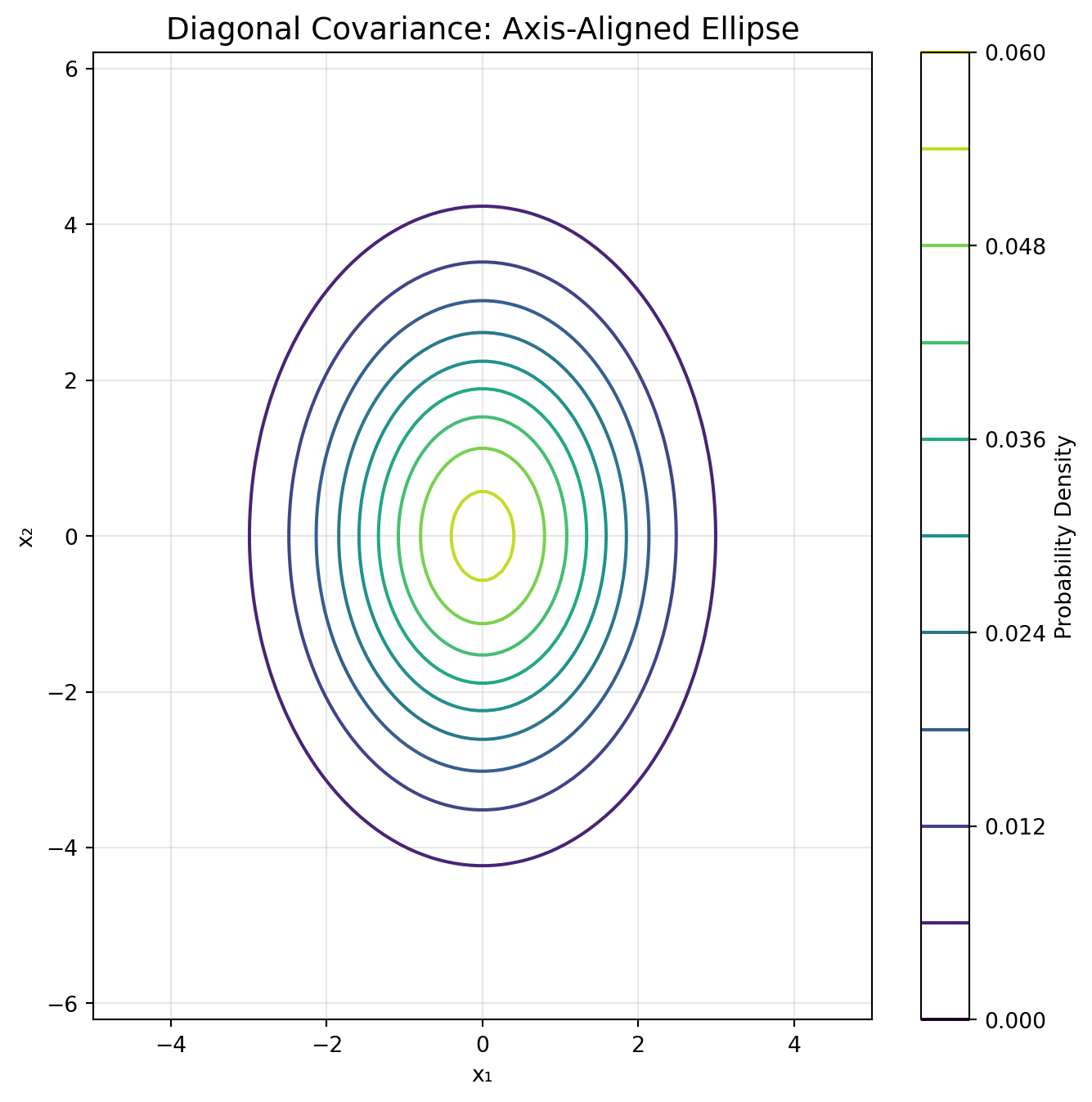
# Cluster 2: Diagonal (axis-aligned ellipse, wider in y)
n2 = 167
X2 = np.random.multivariate_normal([-2, 1], [[0.5, 0], [0, 2.0]], n2)
y2 = np.ones(n2)
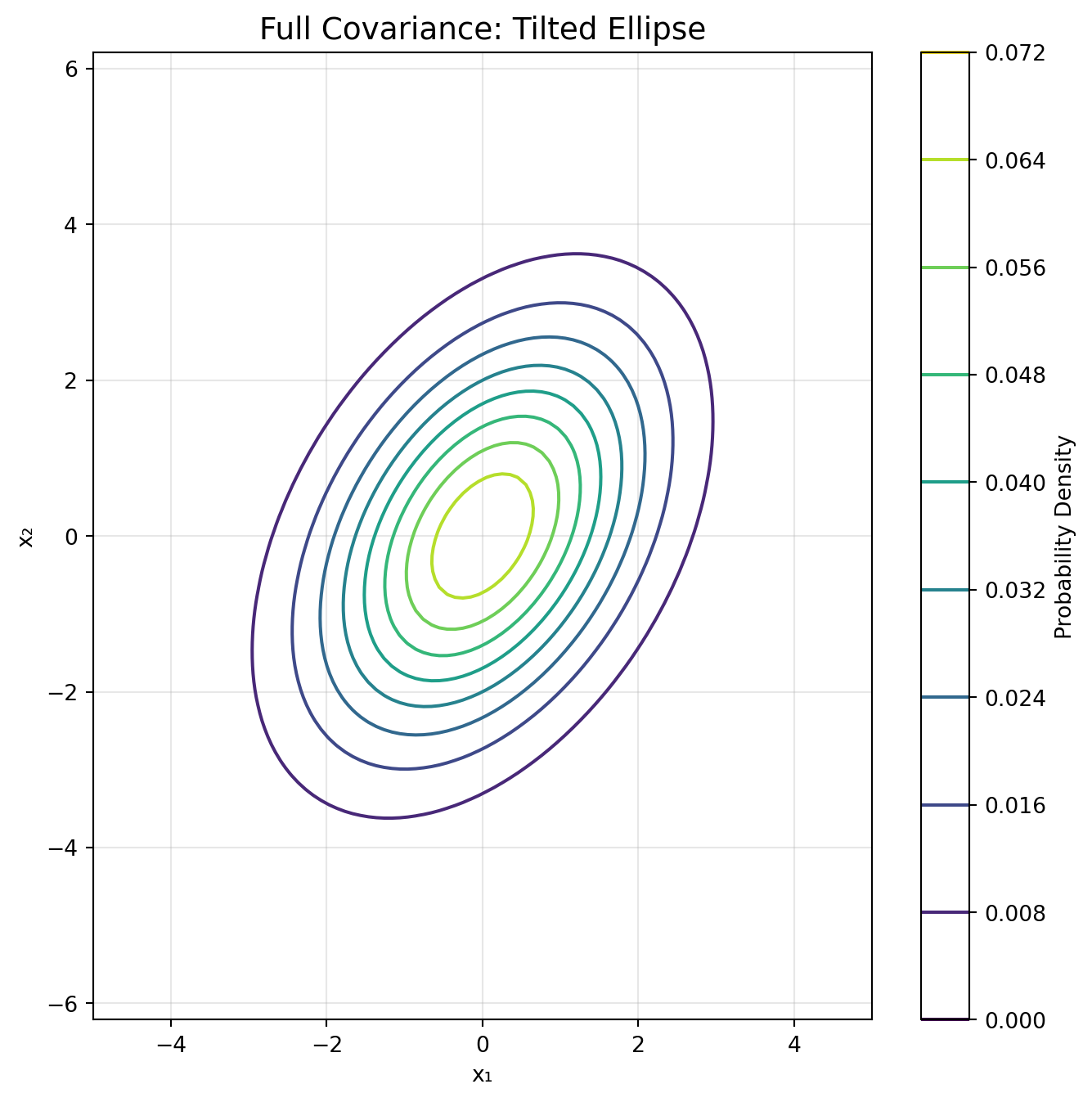
# Cluster 3: Full covariance (tilted ellipse)
n3 = 166
cov3 = np.array([[1.2, 0.8], [0.8, 0.6]]) # Positive correlation
X3 = np.random.multivariate_normal([0, -2], cov3, n3)
y3 = np.full(n3, 2)
# Combine all clusters
X = np.vstack([X1, X2, X3])
y_true = np.hstack([y1, y2, y3])
# Fit GMMs with different covariance types
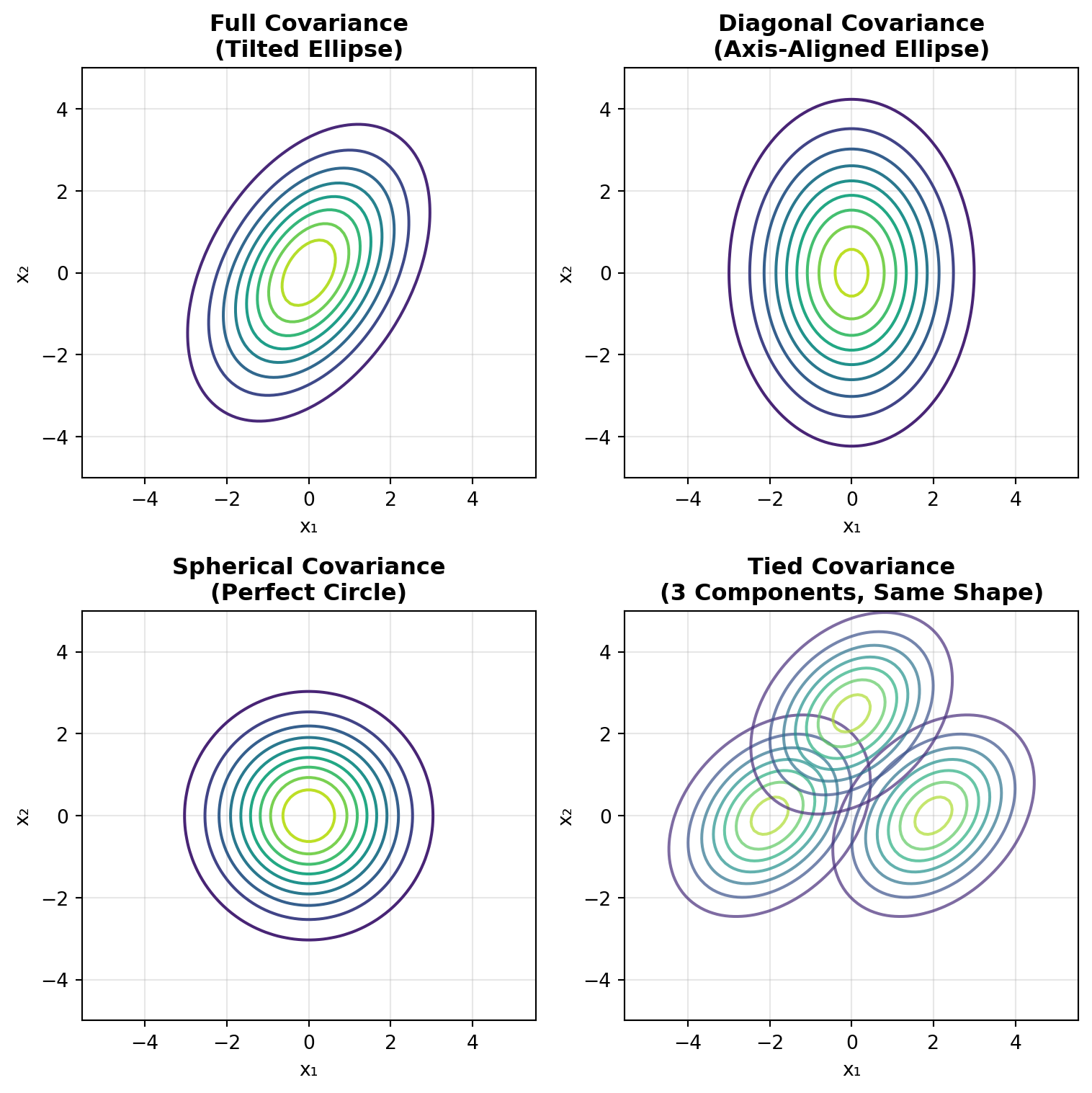
covariance_types = ['full', 'diag', 'spherical', 'tied']
fig, axes = plt.subplots(2, 2, figsize=(8, 8))
for idx, (cov_type, ax) in enumerate(zip(covariance_types, axes.ravel())):
# Fit GMM
gmm = GaussianMixture(n_components=3, covariance_type=cov_type, random_state=42)
gmm.fit(X)
labels = gmm.predict(X)
# Plot data points colored by cluster
scatter = ax.scatter(X[:, 0], X[:, 1], c=labels, s=20, cmap='viridis', alpha=0.6)
# Plot cluster centers
centers = gmm.means_
ax.scatter(centers[:, 0], centers[:, 1], c='red', s=200, alpha=0.8,
marker='X', edgecolors='black', linewidths=2, label='Centers')
# Draw confidence ellipses for each component
from matplotlib.patches import Ellipse
import matplotlib.transforms as transforms
for i in range(3):
if cov_type == 'full':
covariance = gmm.covariances_[i]
elif cov_type == 'diag':
covariance = np.diag(gmm.covariances_[i])
elif cov_type == 'spherical':
covariance = gmm.covariances_[i] * np.eye(2)
elif cov_type == 'tied':
covariance = gmm.covariances_
# Calculate eigenvalues and eigenvectors
v, w = np.linalg.eigh(covariance)
v = 2.0 * np.sqrt(2.0) * np.sqrt(v) # 95% confidence
angle = np.degrees(np.arctan2(w[1, 0], w[0, 0]))
# Draw ellipse
ell = Ellipse(centers[i], v[0], v[1], angle=angle,
edgecolor='red', facecolor='none', linewidth=2, linestyle='--')
ax.add_patch(ell)
ax.set_title(f'{cov_type.capitalize()} Covariance', fontsize=14, fontweight='bold')
ax.set_xlabel('Feature 1')
ax.set_ylabel('Feature 2')
ax.set_xlim([-4, 6])
ax.set_ylim([-4, 6])
ax.legend()
ax.grid(True, alpha=0.3)
plt.tight_layout()
plt.show()
# Print BIC scores for comparison
print("\nBIC Scores (lower is better):")
for cov_type in covariance_types:
gmm = GaussianMixture(n_components=3, covariance_type=cov_type, random_state=42)
gmm.fit(X)
print(f" {cov_type.capitalize()}: {gmm.bic(X):.2f}")